Mapping and analysis of phosphorylation
Protein kinases play a crucial role in the regulation of many cellular processes. They alter the functions of their target proteins by phosphorylating specific serine, threonine and tyrosine residues.
Identification of phosphorylation site sequences and studies with corresponding model peptides have provided clues to how these important enzymes recognize their substrate proteins. This knowledge has made it possible to identify potential sites of phosphorylation in newly sequenced proteins as well as to construct specific model substrates and inhibitors.
One of the most difficult and important challenges in cell biology and medicine is to understand how signaling networks integrate and relay signals. An important step in analyzing signaling networks is the identification and characterization of phosphorylation sites, both on individual proteins and more globally on the proteome.
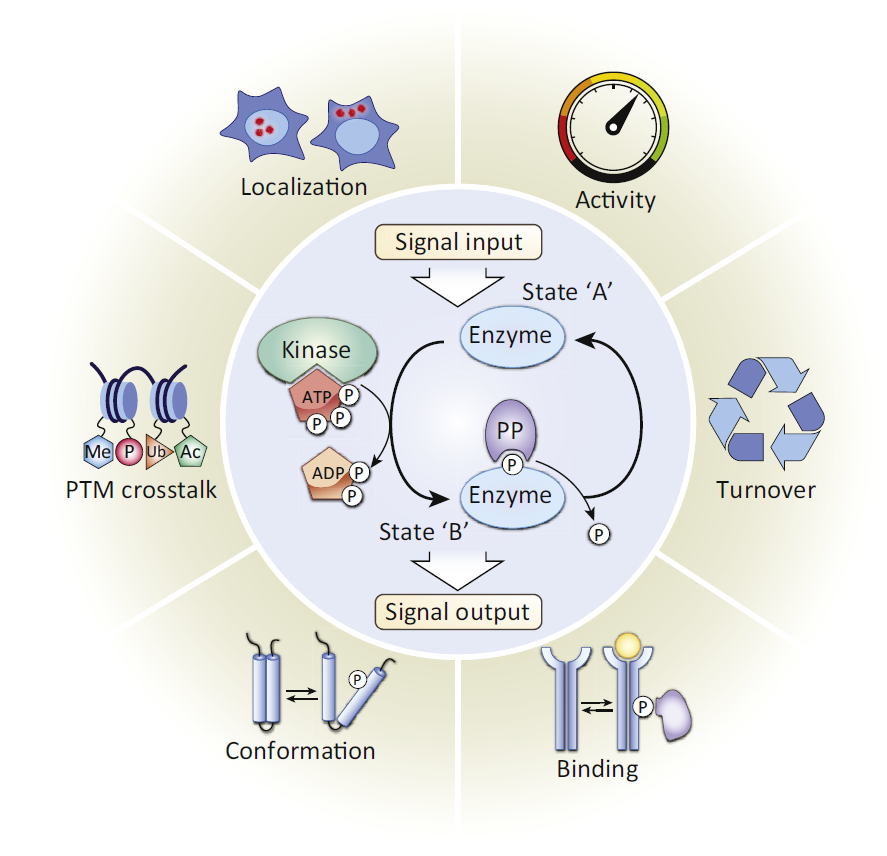
When done carefully, phosphorylation-site analysis provides definitive information on functional relationships between signaling proteins. In the case of multisite phosphorylation on a single protein, it is also a necessary step in defining the mechanism of phosphorylation (i.e., processive or distributive), which determines the nature of the signaling response.
Although protein phosphorylation has been studied for decades, recent advances in mass spectrometry (MS) have revolutionized analysis of signaling by allowing rapid identification of phosphorylation sites with precision and sensitivity. Nevertheless, limitations remain that have important implications for the design of mapping experiments and the subsequent interpretation of biological experiments that make use of the MS-derived data.
Here we provide an overview of technical considerations for successful analysis of serine, threonine, and tyrosine phosphorylation events by mass spectrometry, as well as a discussion of alternative, “old-fashioned” approaches that can be used to complement and/or bolster mass spectrometry data. We have not aimed to provide comprehensive protocols.
