GPS 5.0: An Update on the Prediction of Kinase
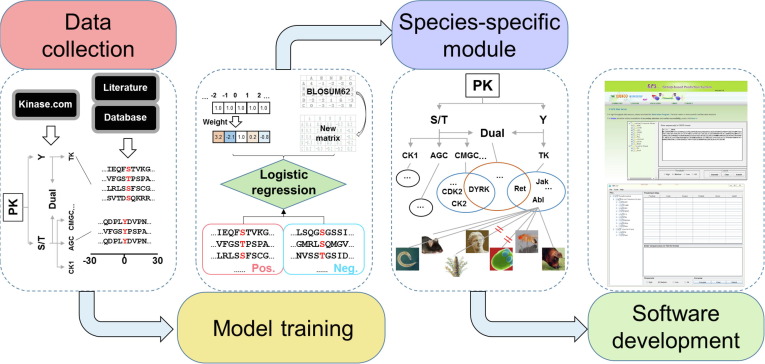
In eukaryotes, protein phosphorylation is specifically catalyzed by numerous protein kinases (PKs), faithfully orchestrates various biological processes, and reversibly determines cellular dynamics and plasticity. Here we report an updated algorithm of Group-based Prediction System (GPS) 5.0 to improve the performance for predicting kinase-specific phosphorylation sites (p-sites).
Two novel methods, position weight determination (PWD) and scoring matrix optimization (SMO), were developed. Compared with other existing tools, GPS 5.0 exhibits a highly competitive accuracy. Besides serine/threonine or tyrosine kinases, GPS 5.0 also supports the prediction of dual-specificity kinase-specific p-sites.
In the classical module of GPS 5.0, 617 individual predictors were constructed for predicting p-sites of 479 human PKs. To extend the application of GPS 5.0, a species-specific module was implemented to predict kinase-specific p-sites for 44,795 PKs in 161 eukaryotes.
Protein phosphorylation plays a critical role in almost all of biological processes and greatly expands the proteome diversity. By covalently attaching phosphate moieties to serine, threonine, and/or tyrosine residues in a dynamic manner, phosphorylation can reversibly change the structure, enzymatic activity, and subcellular trafficking of proteins [1], [2].
In eukaryotes, phosphorylation reaction is differentially and specifically catalyzed by numerous protein kinases (PKs), and each PK only modifies a limited subset of substrates to ensure the signaling fidelity [3], [4], [5].
Aberrances in either PKs or phosphorylated substrates are highly associated with human diseases such as cancer [6], [7]. Therefore, the identification of kinase-specific phosphorylation sites (p-sites) is fundamental for understanding the regulatory mechanisms of phosphorylation.
