KinasePhos: a web tool for identifying protein
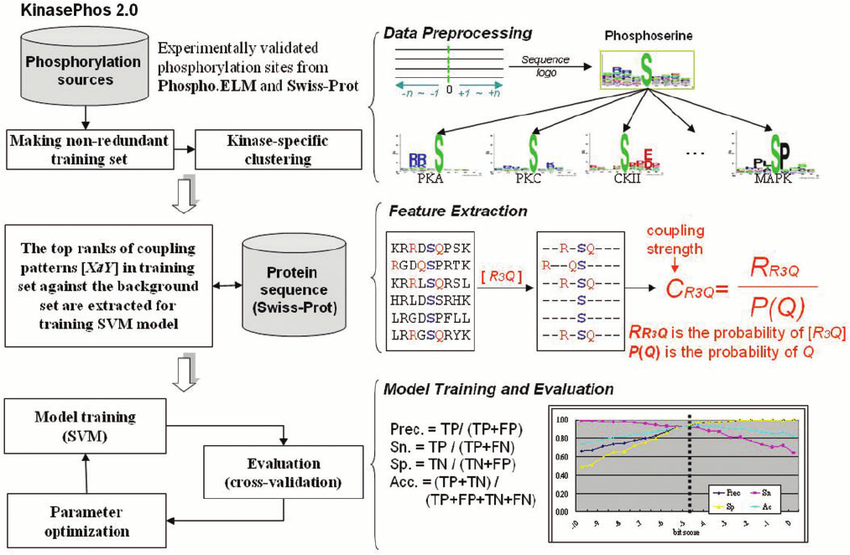
KinasePhos is a novel web server for computationally identifying catalytic kinase-specific phosphorylation sites. The known phosphorylation sites from public domain data sources are categorized by their annotated protein kinases. Based on the profile hidden Markov model, computational models are learned from the kinase-specific groups of the phosphorylation sites.
After evaluating the learned models, the model with highest accuracy was selected from each kinase-specific group, for use in a web-based prediction tool for identifying protein phosphorylation sites. Therefore, this work developed a kinase-specific phosphorylation site prediction tool with both high sensitivity and specificity.
Protein phosphorylation, performed by a group of enzymes known as kinases and phosphotransferases, is a post-translational modification essential to correct functioning within cells ( 1 ).
The post-translational modification of proteins by phosphorylation is the most abundant form of cellular regulation, affecting many cellular signal pathways, including metabolism, growth, differentiation and membrane transport ( 2 ). The enzymes must be specific and act only on a defined subset of cellular targets to ensure signal fidelity.
