Kinase inhibition profiles as a tool to identify kinases
There are thousands of known cellular phosphorylation sites, but the paucity of ways to identify kinases for particular phosphorylation events remains a major roadblock for understanding kinase signaling.
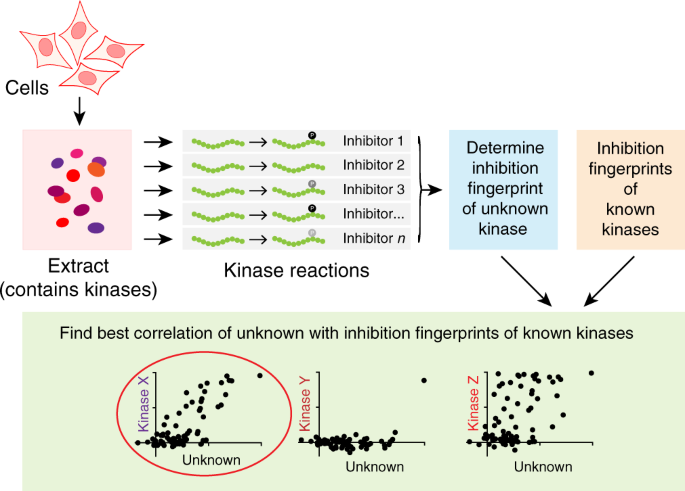
To address this, we here develop a generally applicable method that exploits the large number of kinase inhibitors that have been profiled on near-kinome-wide panels of protein kinases. The inhibition profile for each kinase provides a fingerprint that allows identification of unknown kinases acting on target phosphosites in cell extracts.
We validate the method on diverse known kinase-phosphosite pairs, including histone kinases, EGFR autophosphorylation, and Integrin β1 phosphorylation by Src-family kinases. We also use our approach to identify the previously unknown kinases responsible for phosphorylation of INCENP at a site within a commonly phosphorylated motif in mitosis (a non-canonical target of Cyclin B-Cdk1), and of BCL9L at S915 (PKA). We show that the method has clear advantages over in silico and genetic screening.
Protein phosphorylation is a vital regulatory system in cells, with 90% of all proteins undergoing phosphorylation1. Advances in phosphoproteomics mean that tens of thousands of phosphorylation sites are now known2,3. Large catalogues of specific phosphorylation events that occur within particular cell types or organelles, or during specific treatments or phases of the cell cycle, have been generated4,5,6,7,8.
However, understanding the biology controlled by a particular phosphorylation event depends on being able to identify the kinase(s) responsible for catalyzing it. Unfortunately, the methods available for assigning kinase-phosphosite dependencies are limited. Consequently, our ability to benefit from the expanding knowledge of cellular phosphorylation is hampered.
Of the techniques that have been developed to interrogate kinase-phosphosite dependencies, most focus on identifying the targets of a kinase of interest; far fewer perform the similarly valuable task of identifying the kinase(s) that phosphorylate a particular residue within a substrate9.
Indeed, we are not aware of an experimental method that has found widespread use that can selectively identify direct upstream kinases for any substrate. Broadly, existing approaches can be divided into three categories: (i) in silico predictions; (ii) screens in intact cells, and (iii) biochemical methods, often using cell extracts or recombinant kinases.
