The purpose of this work is to enhance KinasePhos
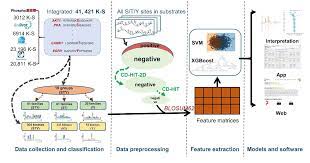
The purpose of this work is to enhance KinasePhos, a machine-learning-based kinase-specific phosphorylation site prediction tool. Experimentally verified kinase-specific phosphorylation data were collected from PhosphoSitePlus, UniProt, GPS 5.0, and Phospho.ELM. In total, 41,421 experimentally verified kinase-specific phosphorylation sites were identified.
A total of 1380 unique kinases were identified, including 753 with existing classification information from KinBase and the remaining 627 annotated by building a phylogenetic tree. Based on this kinase classification, a total of 771 predictive models were built at the individual, family, and group levels, using at least 15 experimentally verified substrate sites in positive training datasets. The improved models were observed to be more effective than other prediction tools.
For example, the prediction of sites phosphorylated by the Akt, CKT, and PKA families had accuracies of 94.5%, 92.5%, and 90.0%, respectively. The average prediction accuracy for all 771 models was 87.2%. For enhancing interpretability, the Shapley additive explanations (SHAP) method was employed to assess feature importance. The web interface of KinasePhos 3.0 has been redesigned with the goal of providing comprehensive annotations of kinase-specific phosphorylation sites on multiple proteins.
Protein phosphorylation is an important eukaryotic post-translational modification. It involves the transfer of a phosphate group from ATP to specific amino-acid residues in the substrate. Phosphorylation is catalyzed by a number of protein kinases, which regulate a variety of signaling pathways and biological functions important in DNA repair, transcriptional regulation, apoptosis, immune response, signaling, metabolism, proliferation, and differentiation. Dysregulation of intracellular phosphorylation networks contributes to the occurrence and development of multiple multifactorial diseases, including cancer, cardiovascular disease, obesity, and others.
Therefore, regulating phosphorylation networks by mediating kinase activity has become an attractive therapeutic strategy with kinases being one of the most important drug targets. Thus, linking dysregulated phosphorylation sites to candidate kinase targets is critical, both for the study of disease mechanisms and the development of therapeutic kinase inhibitors
