Deciphering Protein Kinase
Kinases are master regulators of cellular behavior. Because of the large number of kinases and the even larger number of substrates, approaches that permit global analysis are valuable tools for investigating kinase biology. Mok et al. identified the phosphorylation site selectivity for 61 of the 122 kinases in Saccharomyces cerevisiae by screening a miniaturized peptide library.
By integrating these data with other data sets and structural information, they revealed information about the relationship between kinase catalytic residues and substrate selectivity. They also identified and experimentally verified substrates for kinases, including one for which limited functional information was previously available, showing the potential for this type of analysis as a launching point for the exploration of the biological functions of kinases.
Phosphorylation is a universal mechanism for regulating cell behavior in eukaryotes. Although protein kinases target short linear sequence motifs on their substrates, the rules for kinase substrate recognition are not completely understood. We used a rapid peptide screening approach to determine consensus phosphorylation site motifs targeted by 61 of the 122 kinases in Saccharomyces cerevisiae.
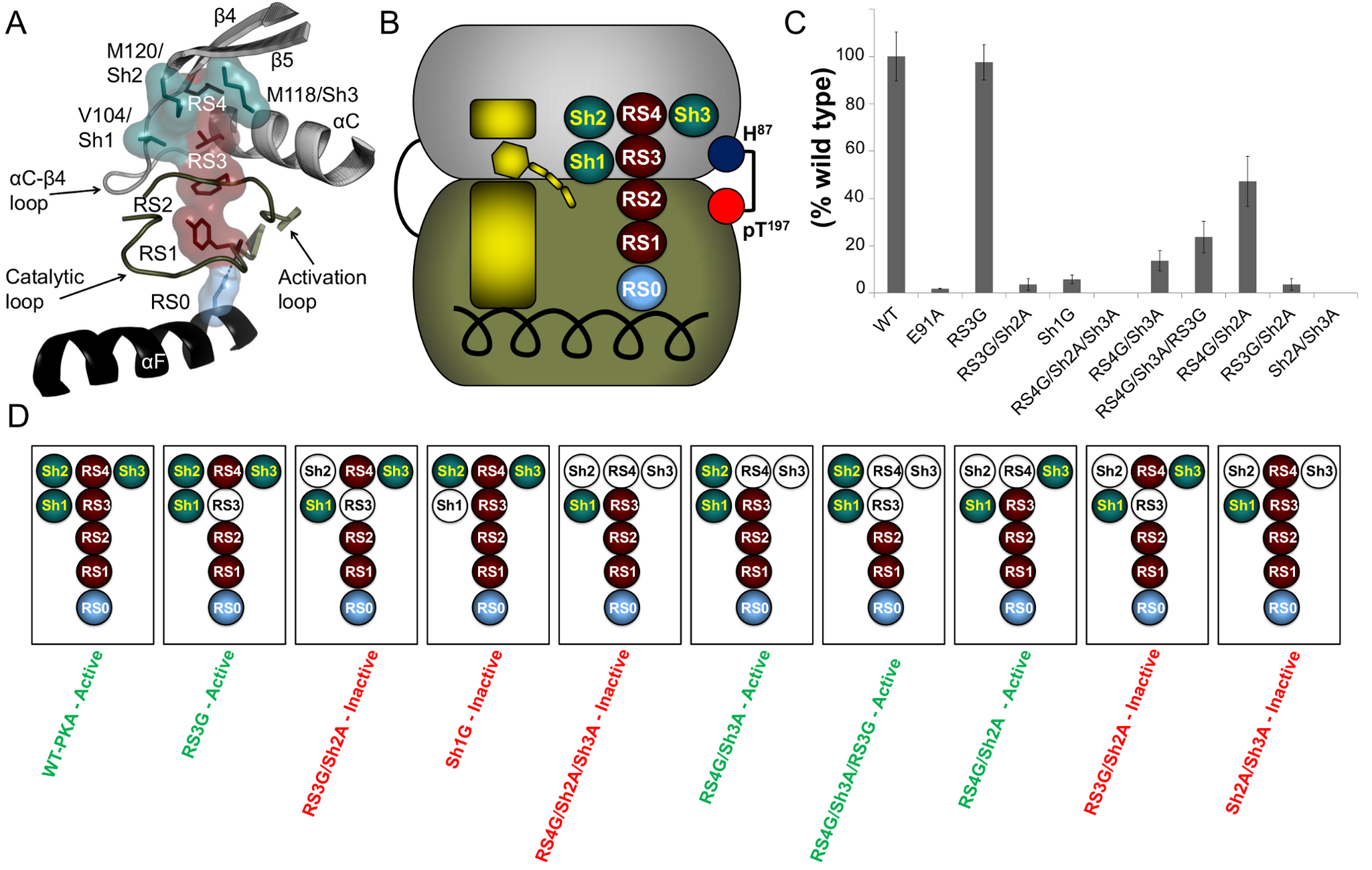
By correlating these motifs with kinase primary sequence, we uncovered previously unappreciated rules for determining specificity within the kinase family, including a residue determining P−3 arginine specificity among members of the CMGC [CDK (cyclin-dependent kinase), MAPK (mitogen-activated protein kinase), GSK (glycogen synthase kinase), and CDK-like] group of kinases. Furthermore, computational scanning of the yeast proteome enabled the prediction of thousands of new kinase-substrate relationships.
We experimentally verified several candidate substrates of the Prk1 family of kinases in vitro and in vivo and identified a protein substrate of the kinase Vhs1. Together, these results elucidate how kinase catalytic domains recognize their phosphorylation targets and suggest general avenues for the identification of previously unknown kinase substrates across eukaryotes.
