Predicting catalyzing kinases of phosphorylation
Protein phosphorylation by kinases plays crucial roles in various biological processes including signal transduction and tumorigenesis, thus a better understanding of protein phosphorylation events in cells is fundamental for studying protein functions and designing drugs to treat diseases caused by the malfunction of phosphorylation.
Although a large number of phosphorylation sites in proteins have been identified using high-throughput phosphoproteomic technologies, their specific catalyzing kinases remain largely unknown. Therefore, computational methods are urgently needed to predict the kinases that catalyze the phosphorylation of these sites.
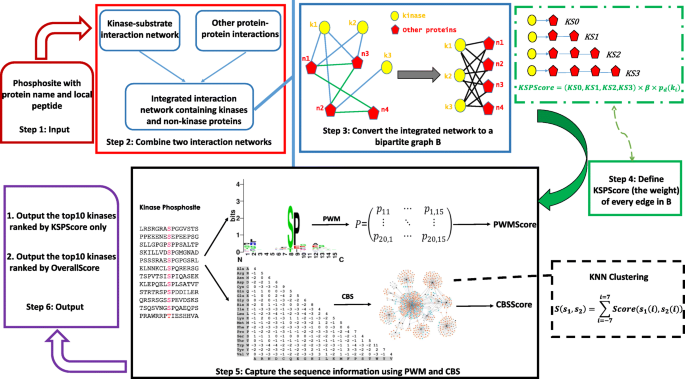
We developed KSP, a new algorithm for predicting catalyzing kinases for experimentally identified phosphorylation sites in human proteins.
KSP constructs a network based on known protein-protein interactions and kinase-substrate relationships. Based on the network, it computes an affinity score between a phosphorylation site and kinases, and returns the top-ranked kinases of the score as candidate catalyzing kinases.
When tested on known kinase-substrate pairs, KSP outperforms existing methods including NetworKIN, iGPS, and PKIS.
Protein kinase selectivity is largely governed by direct binding to the target site(s) on the substrate. Thus, substrate determinants identified from sequences around phosphorylation sites are desirable resources for matching kinases to their substrates. In this study, we tried to identify kinase-selective substrate determinants, including motif sequences, based on large-scale discovery of kinase/substrate pairs. For this purpose, we employed a combination strategy of in vitro kinase reaction followed by LC–MS/MS analysis and applied it to three well-studied kinases: c-AMP regulated protein kinase A (PKA), extracellular signal-regulated kinase 1 (ERK1), and RAC-alpha serine/threonine-protein kinase (AKT1). Cellular proteins were fractionated, dephosphorylated with thermosensitive alkaline phosphatase, phosphorylated with the target kinase, and digested with Lys-C/trypsin, and then phosphopeptides were enriched using TiO2-based hydroxy acid-modified metal oxide chromatography (HAMMOC) and subjected to LC–MS/MS. As a result, 3585, 4347, and 1778 in vitro phosphorylation sites were identified for PKA, ERK1, and AKT1, respectively. As expected, these extensive identifications of phosphorylation sites enabled extraction of both known and novel motif sequences, and this in turn permitted fine discrimination of the specificities of PKA and AKT1, which both belong to the AGC kinase family. Other unique features of the kinases were also characterized, including phospho-acceptor preference (Ser or Thr) and bias ratio of singly/multiply phosphorylated peptides. More motifs were found with this methodology as compared with target kinase phosphorylation of peptides obtained by predigestion of proteins with Lys-C/trypsin. Thus, this approach to characterization of kinase substrate determinants is effective for identification of kinases associated with particular phosphorylation sites.
